In the nucleus of eukaryotic cells, DNA molecules are tightly packed in the form of chromatin, made up of elementary units, the nucleosomes, themselves formed by a long segment of DNA wrapped around a core of 8 histone proteins. Nucleosomes control DNA accessibility by associating and dissociating along genomes and, in so doing, are directly involved in most nuclear processes, such as RNA transcription and DNA repair. Today, it's widely accepted that nucleosomes are organized at active regions (genes, promoters, enhancers) and randomly distributed throughout the rest of the genome. Despite intense efforts to map their positioning using next-generation sequencing (NGS) data, the mechanism(s) behind their collective organization along the genome remains poorly understood.
In this study, the researchers analyzed the genomic positioning profiles of all the nucleosomes covering each coding gene (transcribed regions) of the yeast Saccharomyces cerevisiae, classifying these genes according to their similarities and differences using a correlation coefficient between the nucleosome positioning profiles along each gene. The result of this classification was represented in a space created from a functional principal component analysis of the same profiles. The advantage of this analysis strategy, compared with existing methodologies, is that it takes into account both the details of individual nucleosome positioning, and the overall effect of each nucleosome spanning a gene. Nucleosome distribution was studied in 16 yeast strains, one wild-type and the others deficient in various chromatin remodeler complexes, some of which are known to be involved in the formation of Nucleosome Depleted Regions (NDRs), favorable for transcription initiation (see Joliot news). The strains were compared by decomposing the NGS signals into their main descriptive functions, while retaining the position-specific details of each nucleosome, and considering the arrangement of nucleosomes as a whole. By integrating other genomic properties into their analysis, such as gene size and nucleosome depleted region (NDR) length, the authors identified key factors influencing nucleosome distribution. In particular, they show that Remodels the Structure of Chromatin (RSC) is essential for uncoupling the regular arrangement of nucleosomes within a gene, compared with their disordered structure in inter-gene regions. They also observe that, in a strain deficient in the chromatin remodeling protein Chd1, nucleosome positioning correlates with the presence of RNA polymerase II.
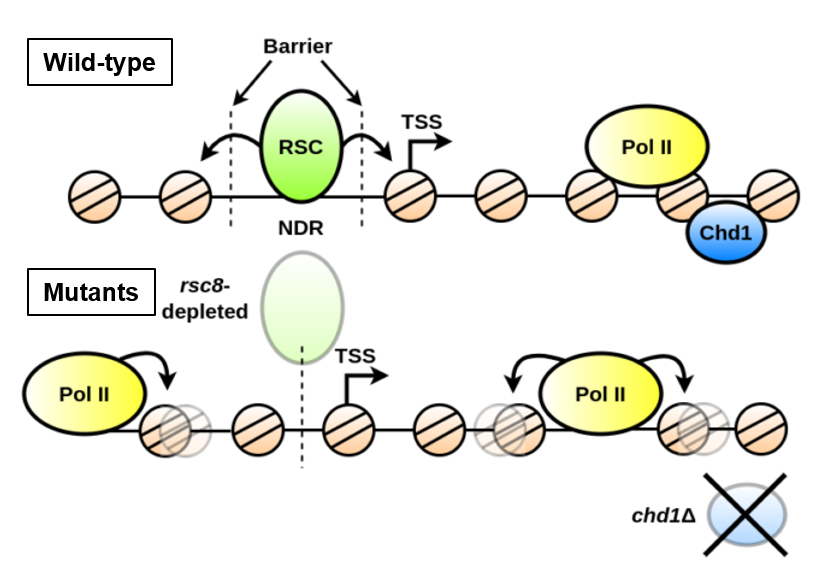
Taken together, these results suggest that chromatin remodelers organize nucleosomes i) at a global level, which defines coding zones where the individual positioning of each nucleosome is modulated by the distribution of other nucleosomes in that zone, ii) at a local level, which tends to individualize the positioning of each nucleosome. The balance between these two levels of organization is necessary to maintain genome integrity and the proper functioning of nuclear processes.
Contact: Arach Goldar arach.goldar@i2bc.paris-saclay.fr