In a cell, there is a whole host of proteins that work around DNA to read it, protect it and repair it... Among the various players involved in this work, nucleases, which cut DNA, are particularly important for repairing breaks. The human nuclease MRN, formed by the association of two proteins, Mre11 and Rad50, is a key player in homologous recombination and meiosis, two processes that use break repair mechanisms.
A locked nuclease
MRN is not mammalian specific. It is conserved and can be found, for example, in bacteria (under the gentle name of SbcC-SbcD). In eukaryotes, the nuclease is equipped with a lock that controls the activation of the enzyme: this is only possible during certain phases of the cell cycle (entry into G2, when the DNA is duplicated to allow the recombination process) when the lock is phosphorylated.
This lock is CtIP, Sae2 in the yeast Saccharomyces cerevisiae. It is a protein with a largely disordered structure, like a long spaghetti. Molecular objects such as these are real puzzles: their three-dimensional structure is very difficult to determine (too flexible, too many possible configurations, too many parasitic phenomena such as their oligomerisation during crystallisation attempts). In this case, although Sae2 was discovered almost 30 years ago, no one has yet been able to determine its structure.
A structural model confirmed by in vitro and in vivo experiments
In a study published in Molecular Cell, teams from the I2BC, the Institut Curie and the IRB (Bellinzona, Switzerland) unravel the mechanisms of action of Sae2 on MRN.
The AMIG (Molecular Assemblies and Genome Integrity) team at the I2BC has succeeded in modelling the structure of the yeast MRN complex using the AlphaFold2 algorithm. Fascinatingly, the programme hesitates between two states during modelling: inactive (autoinhibited) and active. In particular, the addition of phosphorylated Sae2 favours the active conformation in AlphaFold2's predictions: Sae2 would unlock the MRN complex by establishing a synergistic set of interactions centred on the phosphorylation of a Sae2 serine.
Different sets of mutations in the protein partners were then designed to test the model: some mutations would destabilise the network of interactions as it appears in the models predicted by AlphaFold2, while others would combine several individual mutations that compensate each other. The teams from the Institut Curie and the IRB have carried out experiments in vitro (on mutated and purified recombinant proteins) and in vivo (measuring the rate of recombination of DNA breaks, testing meiotic phenotypes) that validate the structural model obtained with AlphaFold2.
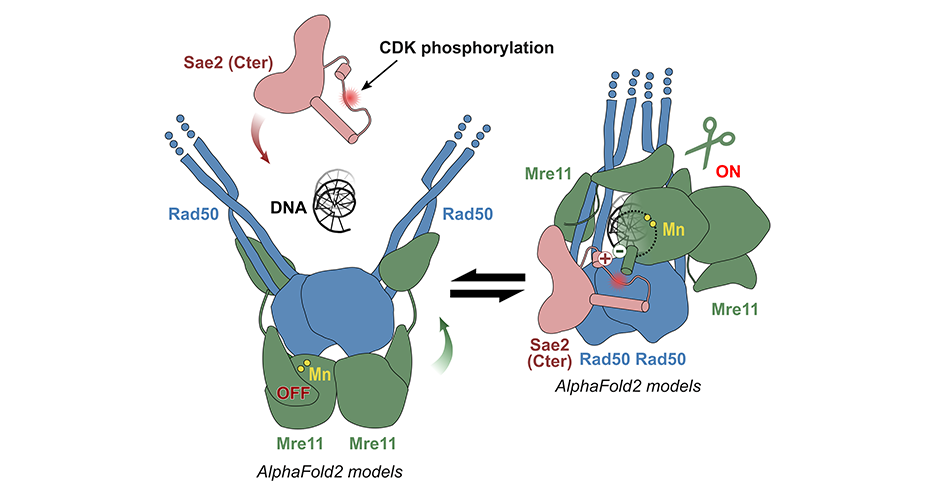
Structural models of the MRN nuclease (Mre11-Rad50) predicted by AlphaFold2: on the left, an inactive model (auto-inhibited) in which Rad50 blocks the active site of Mre11; on the right, an active model favoured by the Sae2 protein phosphorylated in S267. © R. Guerois / CEA
Are the mechanisms described in yeast conserved in humans?
The sequences of Sae2 and CtIP have diverged significantly during evolution. However, in an evolutionary analysis, the researchers show that CtIP, like Sae2, is able to maintain the active conformation of Mre11-Rad50, but with a very different structural motif.
All these data show how an apparently disordered protein can override the auto-inhibition of a nuclease and thus control the switching between different repair pathways, which is important in humans for progression through the cell cycle and meiosis.
Contact within Frédéric-Joliot Institute for Life Sciences / I2BC: