Proteins are important molecules in the living world, as they carry out most biological functions. Their enzymatic modification, in particular the addition or subtraction of various chemical groups, can modulate their activities. This is one of the strategies used by living systems to adapt to the conditions they encounter. In order to monitor these significant changes, a team at IRIG is developing mass spectrometry-based proteomic methods to characterize protein modifications on a large scale.
One of the most frequent enzymatic modifications of proteins involves the addition or substraction of a phosphate group by
protein kinase and phosphatase. These modifications, known as
phosphorylation and dephosphorylation, enable the activity of the target proteins to be finely regulated. It is therefore important to identify which protein sites are modified and the degree of modification, depending on the pathophysiological conditions being studied, in order to decipher the molecular processes involved.
Thanks to its expertise in biochemistry, analytical sciences and data science, an IRIG team has developed a methodology for simultaneously identifying and quantifying thousands of protein phosphorylation sites in complex samples. For example, in the context of studies on Rendu-Osler disease, this method recently led to the discovery of new signalling pathways regulated by BMP9 and BMP10 proteins in endothelial cells (see
Highlight Biosanté) [1]. Another example of the application of this method concerns the understanding of molecular mechanisms used by the hepatitis B virus to hijack the cellular machinery for its own benefit in primary hepatocytes [2].
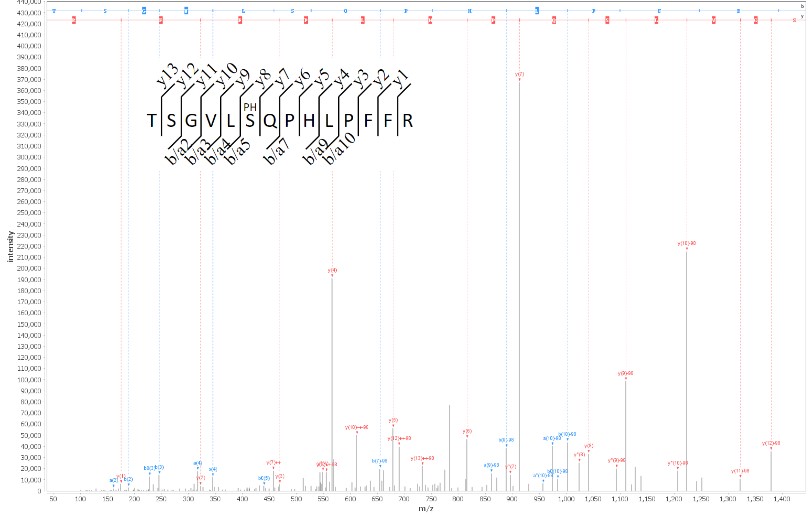
Figure: identification of a phosphorylation site by mass spectrometry-based proteomic analysis. After digesting the proteins in the sample analyzed, the peptides produced are analyzed by mass spectrometry and the spectra generated are interpreted by ad-hoc tools in order to identify their amino acid sequences and modifications.
These developments now make it possible to identify and quantify around 10,000 phosphorylation sites in a complex proteome in a single analysis, providing a better understanding of the mechanisms that regulate the physiological and pathological functioning of biological systems.

Photo: nanoLC-MS coupling (credit CEA)
A
protein kinase is an enzyme that catalyzes the transfer of a phosphate group from a donor molecule to a target protein.
Phosphorylation is the addition of a phosphate group to a protein or small molecule.
Fundings
ANR (Agence Nationale de la Recherche):
- ProFI (Proteomics French Infrastructure)
- GRAL
via Chemistry Biology Health Graduate School at University Grenoble Alpes
ANRS (Agence Nationale de Recherche sur le Sida et les hépatites virales):